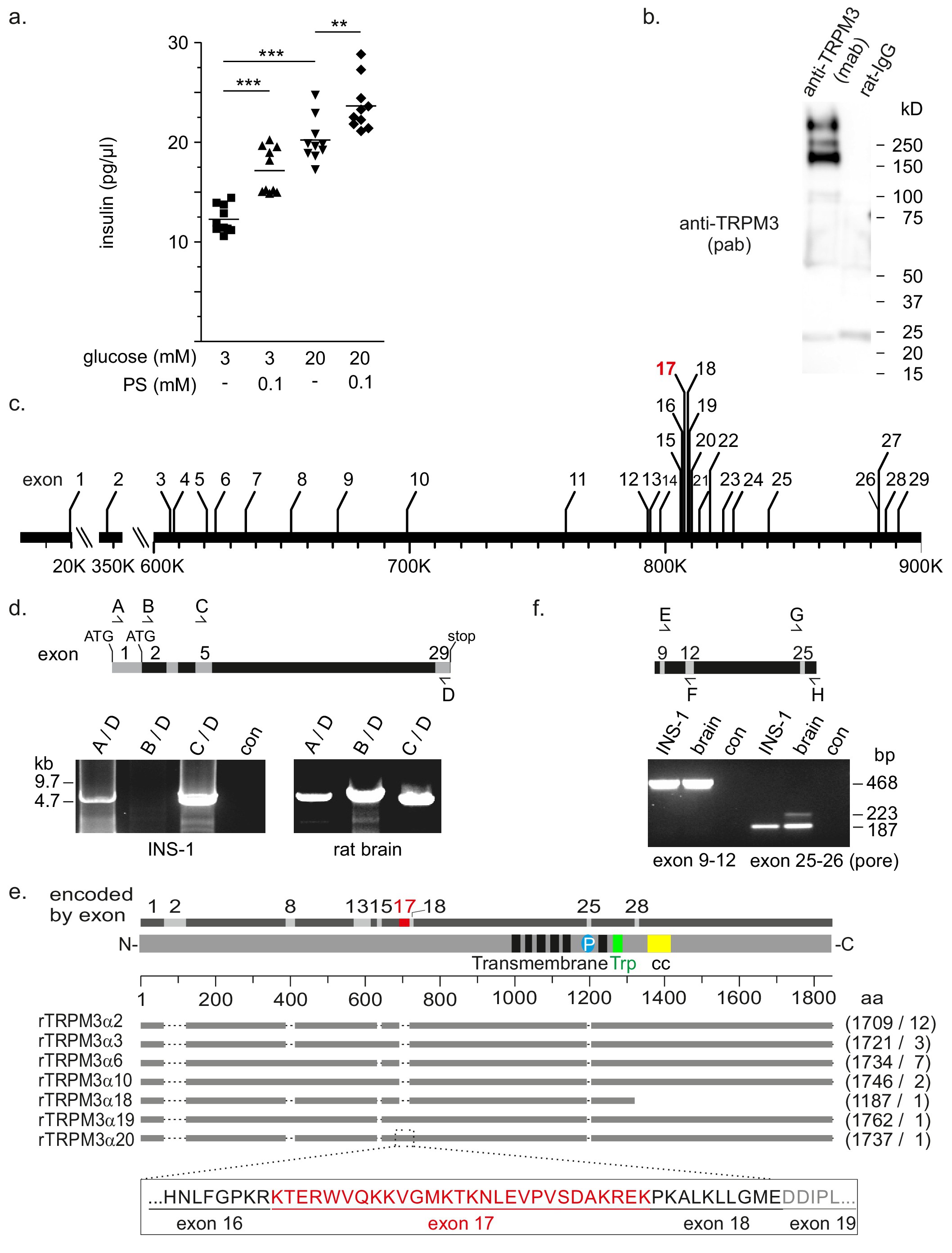
Fig. 1. Properties of INS-1 beta cells as a model to analyse the role of TRPM3 in insulin release. (a) Insulin released from INS-1 cells in the presence of 3 and 20 mM glucose and the absence and presence of 0.1 mM PS. (b) Identification of TRPM3 proteins in INS-1 cells using monoclonal anti-TRPM3-antibodies (mab) from rat for immunoprecipitation and polyclonal anti-TRPM3-antibodies (pab) for detection in a western blot. Rat immunoglobulin G (IgG) served as control. (c) Genomic organization of the rat Trpm3 gene located on chromosome 1q51 spanning ~ 870 kb and comprising 29 exons. A newly identified exon 17 is highlighted in red. (d) Amplification of Trpm3 transcripts by RT-PCR from INS-1 cells and rat brain. The location of the target sequences of the oligonucleotide primers A, B, C, and D within the predicted rat exons (numbers) is indicated in the diagram. Control reactions (con) without template revealed no products. (e) Schematic presentation of TRPM3 protein isoforms (grey bars) identified in INS-1 cells and scaled to their relative size with the numbers of amino acid residues (aa) and the number of identified clones indicated in brackets. Internal protein domains removed by alternative splicing are indicated as dotted lines. The protein region encoded by a newly identified exon 17 (red) is presented below. The organization of TRPM3 protein domains is shown above, with the transmembrane region including the six transmembrane domains (black rectangles), the channel pore (P), the TRP motif (Trp), and a coiled-coil region (cc) as indicated. Alternative protein-coding exons identified in mouse [24] and rat are indicated on top, with the newly identified exon 17 highlighted in red. (f) Detection of pore-coding Trpm3 transcripts in INS-1 cells and rat brain by RT-PCR. The locations of the targets of the oligonucleotide primers G and H to amplify transcripts encoding the pore regions (exon 25-26) and of oligonucleotide primers E and F to amplify fragments common to all Trpm3 transcripts (exon 9-12) as control are indicated.